- Collections
numpyarrays- Functions
Now that we are comfortable with basic types, collections, numpy arrays, and functions it is time to have a look at object-oriented programming. In programming jargon, an object is an ensemble of data and functionality that make sense together.
In Python, everything is an object and therefore the concepts of object-oriented programming are in extensive use in every Python code. So, why are objects so widespread? Because they are very useful to write re-usable code and hide away a lot of complexity.
Let's suppose, for example, that we want to represent a molecule in a Python program. At the very least, we will need to store some data representing the molecule. In order to represent a particular configuration of acetylene, we could store the element number and the atomic coordinates of the atoms in the molecule:
import numpy as np
elements = np.array([1, 6, 6, 1])
coordinates = np.array([
[-1.06, 0.00, 0.00], # H1
[ 0.00, 0.00, 0.00], # C1
[ 1.20, 0.00, 0.00], # C2
[ 2.26, 0.00, 0.00], # H2
])
Coordinates are stored in a $3 \times N$ array ($N$ being the number of atoms) and each row is assumed to be ordered in the same way as elements are.
This is all good, but we'll need to keep track of the fact that the elements and coordinates vectors represent the same molecule and therefore should be, in most cases, processed together. And what if we want to represent another molecule? To represent a dihydrogen molecule in the same program we will need to do something like the following:
elements_H2 = np.array([1, 1])
coordinates_H2 = np.array([
[0.00, 0.00, 0.00], # H1
[1.00, 0.00, 0.00], # H1
])
This works, but it's ugly. It is also error-prone and probably will not scale well to many molecules.
Can you think of a way of tying together
elementsandcoordinateswithin a single variable with what you learned already?
Using a dictionary to represent acetylene could be a good solution:
acetylene = {
"elements": np.array([1, 6, 6, 1]),
"coordinates": np.array([
[-1.06, 0.00, 0.00], # H1
[ 0.00, 0.00, 0.00], # C1
[ 1.20, 0.00, 0.00], # C2
[ 2.26, 0.00, 0.00], # H2
])
}
We now have a single variable representing the acetylene molecule. All the information is stored within that variable and it can be easily and unequivocally accessed. If we want to see the elements (atomic numbers) of the atoms in acetylene, we can simply print them out:
print(acetylene["elements"])
Do you see where we are heading? Objects allow us to tie together data (as we just did with a dictionary) and functionality together. By defining and using objects we can create a super slick representation of a molecule, with all the data and all the functionality we need.
In the following sections we will have a first look at the simple concepts of object-oriented programming (OOP) to get an idea of this powerful paradigm. We'll learn a lot more about OOP in the advanced sections.
A class is the definition of an object. We can think of a class as an architectural plan and an object (usually called an instance of the class in this context) as an house built following said plan. With the same plan, you can build multiple houses (create multiple instances), each housing different people (different data). For our example above, a yet to be defined Molecule class would represent a generic molecule (by defining the data and the functionality associated to a molecule) while acetylene would be an object (instance) of said class, containing the actual data defining acetylene.
Creating a new class in Python has a similar syntax to a function definition, with the class keyword instead of def and without () (() is optional for a class definition):
class EmptyClass:
"""
Empty class that does nothing.
Note:
The `pass` statement in Python does nothing.
"""
pass
Note #1: the pass statement in Python does nothing and it is therefore useful when a statement is required but no code needs to be written.
Note #2: docstrings are used to document classes as well.
Once our class is defined, we can create (instantiate) an object of said class:
ec = EmptyClass()
We can check that the content of the variable ec is indeed an instance of the class EmptyClass using the isinstance function:
isinstance(ec, EmptyClass)
When we create an object from a class definition, we usually need to initialise some data. Let's now see how to do it.
A method is a function associated to a class. Methods are defined in the same way one would define a standard Python function and follow the same rules. However, there are a few names for methods that have special meaning. One of such methods is the __init__ function, which is called to create a new object from a class.
class SimpleMolecule:
"""
Rempresentation of a simple molecular conformation.
Args:
els (numpy.ndarray): array of elements
coords (numpy.dnarray): array of coordinates
Attributes:
elements (numpy.ndarray): array of elements
coordinates (numpy.dnarray): array of coordinates
Note:
The `self` argument is omitted from the documentation.
"""
def __init__(self, els, coords):
self.elements = els
self.coordinates = coords
Note #3: The __init__ method can be documented either at the class level or the __init__ function level. Either form is acceptable, but one needs to be consistent across the code.
Here is a schematic representation of the class SimpleMolecule:
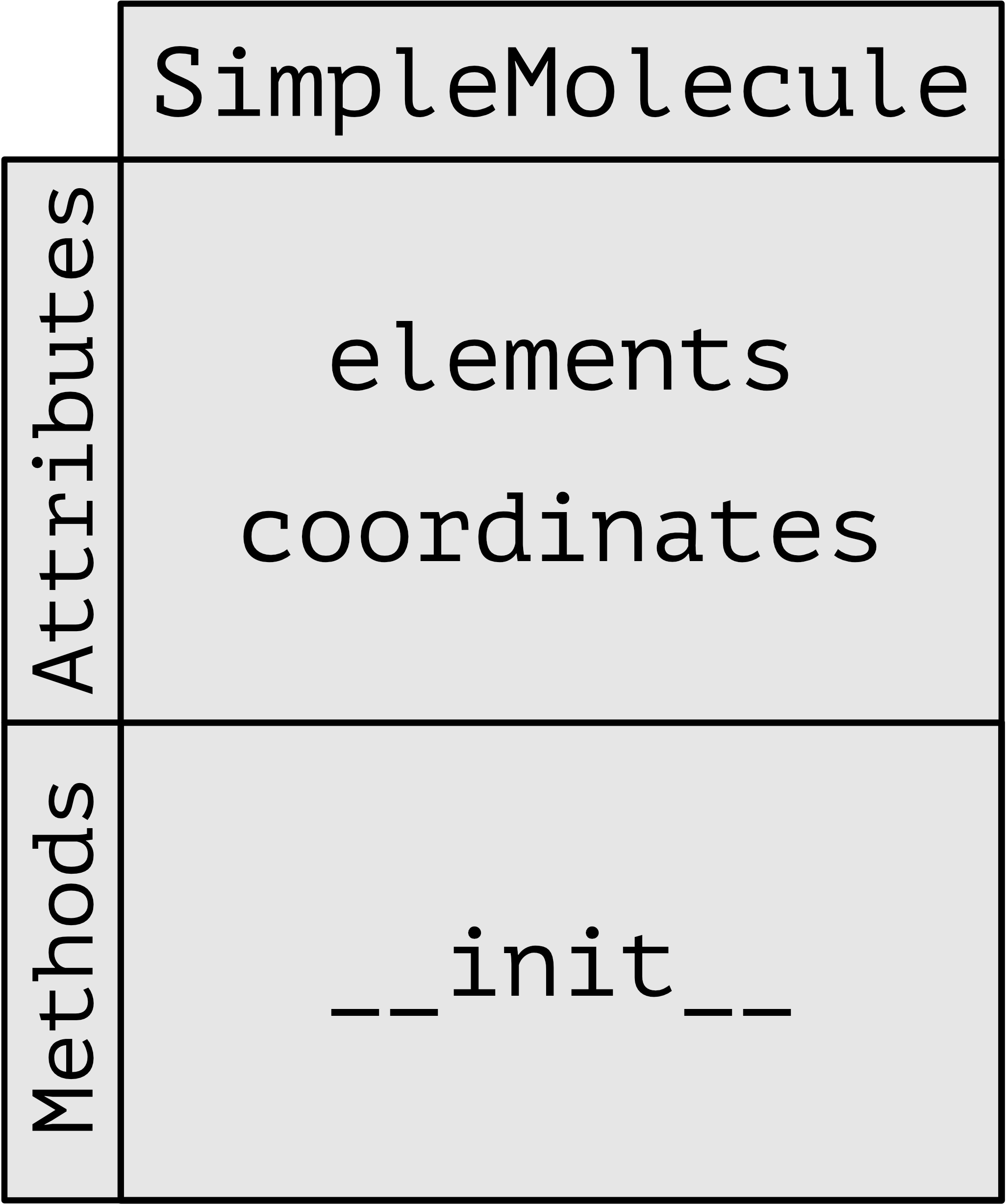
SimpleMolecule is now a class representing a molecule which can be initialised with elements and coordinates. In order to create an acetylene instance of the SimpleMolecule class we can do the following:
acetylene = SimpleMolecule(
els=np.array([1, 6, 6, 1]),
coords=np.array([
[-1.06, 0.00, 0.00], # H1
[ 0.00, 0.00, 0.00], # C1
[ 1.20, 0.00, 0.00], # C2
[ 2.26, 0.00, 0.00], # H2
])
)
There are three important things to notice here. First, the __init__ method takes self as the first of its three arguments. Second, the __init__ function is not called explicitly but it's called implicitly as SimpleMolecule(els, coords); this is why this method is "special" (some Pythonistas would call it magic!). Third, a SimpleMolecule object is created by passing two arguments and not three. The reason behind the latter behavior is that Python always passes, automatically, the object itself (which is an instance of the class SimpleMolecule) as first argument of a method. Therefore, when we initialise SimpleMolecule we only need to define the els and coords arguments, while the self argument is taken care of automatically by Python.
The self argument, which represents the particular instance of the class we are working with, can be used to store data related to that particular instance. The variables stored within a class are called attributes. In the case of SimpleMolecule the attributes are self.elements and self.coordinates. They can be easily accessed with the . operator:
print(acetylene.elements)
Note #4: The acetylene instance of the SimpleMolecule class is very similar to the dictionary we used before. With the dictionary we used print(acetylene["elements"]), while now we use print(acetylene.elements).
In just a few paragraphs we have been through a lot of new and complicated concepts, so let's try to recapitulate the example above. SimpleMolecule is a class which defines how a simple molecule is represented in our Python program. The object acetylene is a particular instance of the SimpleMolecule class, intialised using the __init__ method. acetylene stores the atomic numbers and atomic coordinates in the attributes self.elements and self.coordinates; the values of the attributes are specific to the acetylene instance and they will have different values for different instances.
To verify the latter statement we can create another instance of SimpleMolecule, this time representing dihydrogen:
H2 = SimpleMolecule(
np.array([1, 1]),
np.array([[0.00, 0.00, 0.00], [1.00, 0.00, 0.00]])
)
If we print out the elements and coordinates associated to H2 we see that they are different from the ones associated to acetylene:
print(H2.elements)
print(acetylene.elements)
The SimpleMolecule class specifies the general structure of a simple molecule (as represented in our program), while its instances H2 and acetylene represent actual molecules with the associated data. This example helps clarify the meaning of self: for H2 the attribute self.elements actually means H2.elements while for acetylene the attribute self.elements actually means acetylene.elements.
As outlined in Note #4, the SimpleMolecule class does not differ a lot from the original acetylene dictionary we defined above: it contains the same data which are accessed using acetylene.elements (class) instead of acetylene["elements"] (dictionary). Why we introduced a lot of complicated concepts to accomplish something we can achieve simply with a dictionary? The reason is that with classes we can do much more!
A simple extension of the SimpleMolecule class is to add some functionality. For example, we might want to be able to remove hydrogen atoms. We can add functionality to do that, by defining a method for that purpose:
class Molecule:
"""
Rempresentation of a simple molecular conformation.
Args:
els (numpy.ndarray): array of elements
coords (numpy.dnarray): array of coordinates
Attributes:
elements (numpy.ndarray): array of elements
coordinates (numpy.dnarray): array of coordinates
Note:
The `self` argument is omitted from the documentation.
"""
def __init__(self, els, coords):
self.elements = els
self.coordinates = coords
def removeHs(self):
"""
Remove hydrogen atoms from the molecule.
Note:
Information about hydrogen atoms is lots forever when
this function is called: attributes `elements` and
`coordinates` are modified in-place.
"""
# Mask for hydrogen atoms
# The mask array is True where there are no hydrogen atoms
# The mask array is False where there are hydrogen atoms
mask = self.elements != 1
# Select only elements where the mask is True
self.elements = self.elements[mask]
# Select only coordinates where the mask is True
self.coordinates = self.coordinates[mask, :]
Here is a schematic representation of the class Molecule
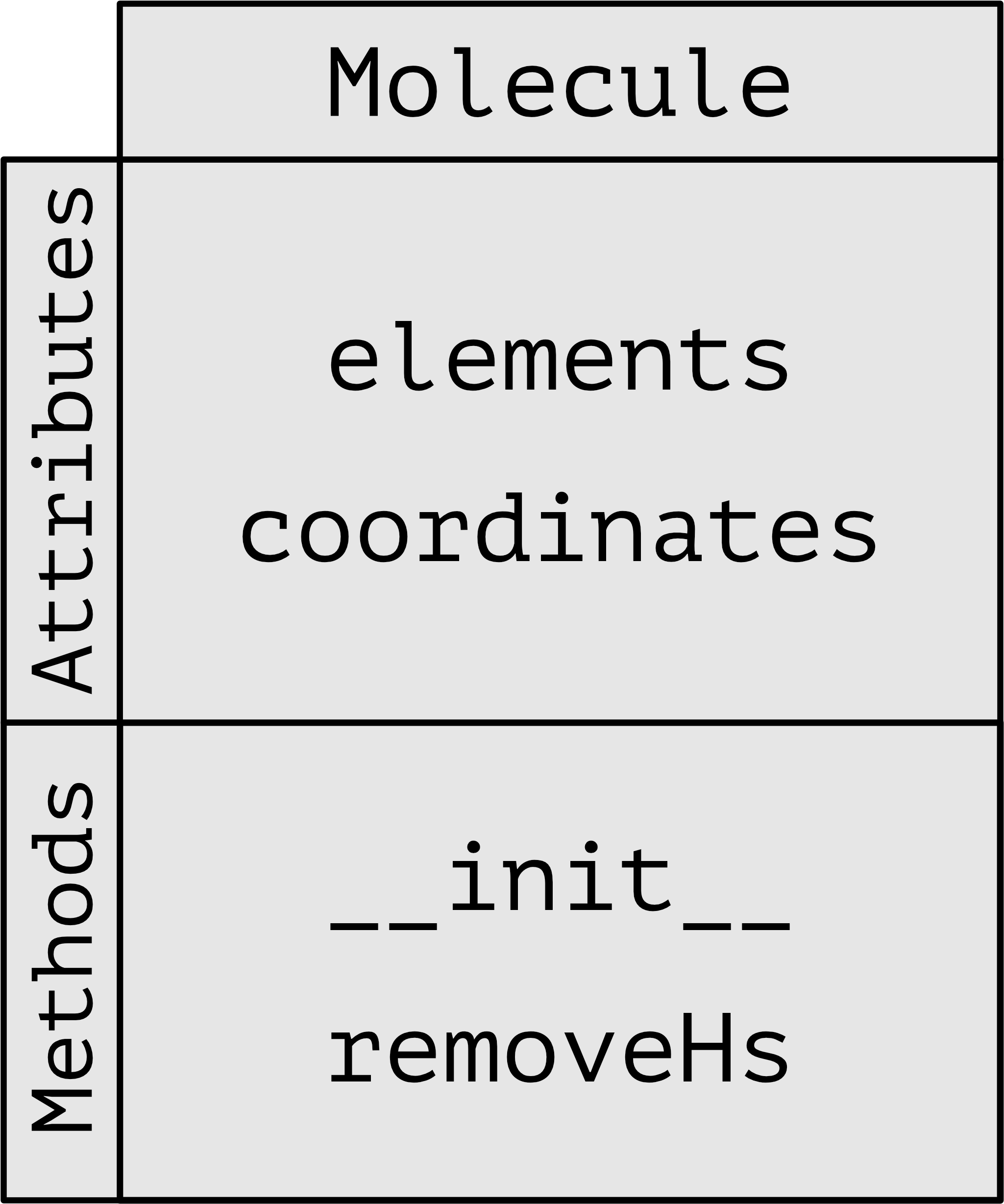
The Molecule class is now more complex and can't be reproduced simply or cleanly with a dictionary. In addition to the self.elements and self.coordinates attributes we now have some functionality (the removeHs method) associated to the Molecule class that allows us to strip all the hydrogens from any molecule with a simple function call. It is important to notice that the method removeHs only takes self as argument since self.elements and self.coordinates, which are attributes, are always available within the class. We can define again an acetylene molecule, but this time as an instance of the Molecule class (and not the SimpleMolecule class we used before):
acetylene = Molecule(
els=np.array([1, 6, 6, 1]),
coords=np.array([
[-1.06, 0.00, 0.00], # H1
[ 0.00, 0.00, 0.00], # C1
[ 1.20, 0.00, 0.00], # C2
[ 2.26, 0.00, 0.00], # H2
])
)
print(acetylene.elements)
The definition of acetylene is the same as before (since the __init__ method did not change), but being an instance of the Molecule class, there is now an associated removeHs method. The method can be called using the . operator and since the self argument of removeH is passed automatically there is no other argument that needs to be specified:
acetylene.removeHs()
If we now print the self.elements and self.coordinates attributes, we can confirm that hydrogen atoms have been removed:
print(acetylene.elements)
print(acetylene.coordinates)
As we can see, after calling removeHs from the acetylene instance of Molecule (using the . operator), the information about hydrogen atoms has been removed from both the self.element and self.coordinates attributes, with a simple function call. Achieving this functionality with the simple dictionary defined above would have been more complicated and error prone.
In this section we have introduced a lot "basic" principles and definitions of object oriented programming and hopefully you are now convinced of how powerful and clean this programming paradigm is. We made extensive use of the . operators, which indicates you are using an object and accessing its data (attributes) or associated functionality (methods). The key points to take away from this lesson are the following:
- In Python, everything is an object
- Objects tie together related data and functionality
- A class is the definition of an object
- The
classkeyword allows us to define a class in Python - An instance of a class is an object of said class which exists in your program as a variable
- The variables associated to an object are called attributes
- We define an attribute by defining a
self.attribute_namevariable within the class
- We define an attribute by defining a
- The functions associated to an object are called methods
- We define a method as a standard Python function, with
selfas first argument
- We define a method as a standard Python function, with
- The
__init__method of a class defines how an instance is initialised
The idea of a molecule API is loosely inspired by Depth-First: A Minimal Molecule API by Richard L. Apodaca
